Restriction Analysis by Southern Blot Analysis
Restriction endonucleases are DNA-cleaving enzymes with defined sequences as targets. They are often simply called restriction enzymes. Since each enzyme cleaves DNA only at its specific recognition sequence, the total DNA of an individual present in nucleated cells can be cut into pieces of manageable and defined size in a reproducible way. Individual DNA fragments can then be selected, ligated into suitable vectors, multiplied, and examined. Owing to the uneven distribution of recognition sites, the DNA fragments differ in size. A starting mixture of DNA fragments is sorted according to size. Two procedures detect target DNA or RNA fragments after they have been arranged by size in gel electrophoresis—the Southern blot hybridization for DNA (named after E. Southern who developed this method 1975) and the Northern blot hybridization for RNA (a word play on Southern, not named after a Dr. Northern). Immunoblotting (Western blot) detects proteins by an antibody-based procedure.
Southern blot hybridization
The analysis starts with total DNA (1). The DNA is isolated and cut with restriction enzymes (2). One of the not yet identified fragments contains the gene being sought or part of the gene. The fragments are sorted by size in a gel (usually agarose) in an electric field (electrophoresis) (3). The smaller the fragment, the faster it migrates; the larger, the slower it migrates. Next, the blot is carried out: The fragments contained in the gel are transferred to a nitrocellulose or nylon membrane (4). There the DNA is denatured (made single-stranded) with alkali and fixed to the membrane by moderate heating (∼ 80°C) or UV cross-linkage. The sample is incubated with a probe of complementary singlestranded DNA (genomic DNA or cDNA) from the gene (5). The probe hybridizes solely with the complementary fragment being sought, and not with others (6). Since the probe is labeled with radioactive 32 P, the fragment being sought can be identified by placing an X-ray film on the membrane, where it appears as a black band on the film after development (autoradiogram) (6). The size, corresponding to position, is determined by running DNA fragments of known size in the electrophoresis.
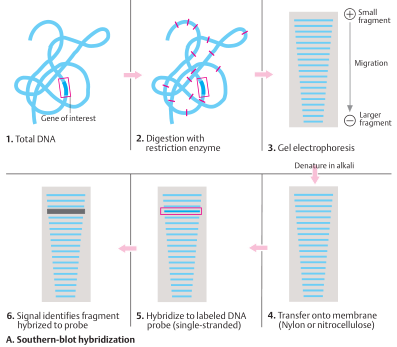
Southern blot hybridization
Restriction fragment length polymorphism (RFLP)
In about every 100 base pairs of a DNA segment, the nucleotide sequence differs in some individuals (DNA polymorphism). As a result, the recognition sequence of a restriction enzyme maybepresent ononechromosome butnotthe other. In this case the restriction fragment sizes differ at this site (restriction fragment length polymorphism, RFLP). An example is shown for two 5 kb (5000 base pair) DNA segments. In one, a restriction site in the middle is present (allele1); in the other (allele2) it is absent. With a Southern blot, it can be determined whether in this location an individual is homozygous 1–1 (two alleles 1, no 5 kb fragment), heterozygous 1–2 (one allele each, 1 and 2), or homozygous 2–2 (two alleles 2). If the mutation being sought lies on the chromosome carrying the 5 kb fragment , the presence of this fragment indicates presence of the mutation. The absence of this fragment would indicate that the mutation is absent. It is important to understand that the RFLP itself is unrelated to the mutation. It simply distinguishes DNA fragments of different sizes from the same region. These can be used as markers to distinguish alleles in a segregation analysis. In addition to RFLPs, other types of DNA polymorphism can be detected by Southern blot hybridization, although polymerase chain reaction-based analysis of microsatellites is now used more frequently.
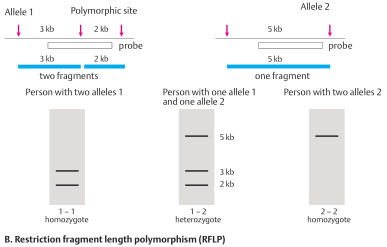
Restriction fragment length polymorphism (RFLP)