Changes in DNA (Mutations)
When it was recognized that changes (mutations) in genes occur spontaneously (T. H. Morgan, 1910) and can be induced by X-rays (H. J. Muller, 1927), the mutation theory of heredity became a cornerstone of early genetics. Genes were defined as mutable units, but the question what genes and mutations are remained. Today we know that mutations are changes in the structure of DNA and their functional consequences. The study of mutations is important for several reasons. Mutations cause diseases, including all forms of cancer. They can be induced by chemicals and by irradiation. Thus, they represent a link between heredity and environment. And without mutations, well-organized forms of life would not have evolved.
The following two plates summarize the chemical nature of mutations.
A. Error in replication
The synthesis of a new strand of DNA occurs by semiconservative replication based on complementary base pairing. Errors in replication occur at a rate of about 1 in 10 5 . This rate is reduced to about 1 in 10 7 to 10 9 by proofreading mechanisms. When an error in replication occurs before the next cell division (here referred to as the first division after the mutation), e.g., a cytosine (C) might be incorporated instead of an adenine (A) at the fifth base pair as shown here, the resulting mismatch will be recognized and eliminated by mismatchre pair in most cases.
However, if the error is undetected and allowed tostand, the next(second) division will result in a mutant molecule containing a CG instead of an AT pair at this position. This mutation will be perpetuated in all daughter cells. Depending on its location within or outside of the coding region of a gene, functional consequences due to a change in a codon could result.
B. Mutagenic alteration of a nucleotide
A mutation may result when a structural change of a nucleotide affects its base-pairing capability. The altered nucleotide is usually present in one strand of the parent molecule. If this leads to incorporation of a wrong base, such as a C instead of a T in the fifth base pair as shown here, the next (second) round of replication will result in two mutant molecules.
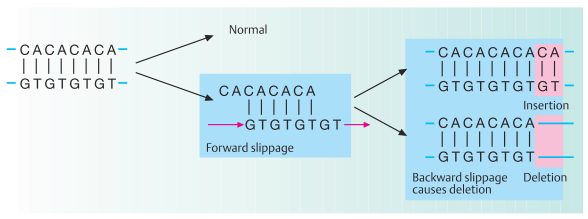
C. Replication slippage
A different class of mutations does not involve an alteration of individual nucleotides, but results from incorrect alignment between allelic or nonallelic DNA sequences during replication. When the template strand contains short tandem repeats, e.g., CA repeats as in microsatellites (see DNA polymorphism and Part II, Genomics), the newly replicated strand and the template strand may shift their positions relative to each other. With replication or polymerase slippage, leading to incorrect pairing of repeats, some repeats are copied twice or not at all, depending on the direction of the shift. One can distinguish forward slippage (shown here) and backward slippage with respect to the newly replicated strand. If the newly synthesized DNA strand slips forward, a region of nonpairing remains in the parental strand. Forward slippage results in an insertion. Backward slippage of the new strand results in deletion. Microsatellite instability is a characteristic feature of hereditary nonpolyposis cancer of the colon (HNPCC). HNPCC genes are localized on human chromosomes at 2p15–22 and 3p21.3. About 15% of all colorectal, gastric, and endometrial carcinomas show microsatellite instability. Replication slippage must be distinguished from unequal crossing-over during meiosis. This is the result of recombination between adjacent, but not allelic, sequences on nonsister chromatids of homologous chromosomes (Figures redrawn from Brown, 1999).
Replication slippage